Scientific reviews | 3
Title: Streamlined Quantitative Imaging Biomarker Development: Generalization of radiomics through automated machine learning
Author: Martijn P. A. Starmans, PhD
Publication: February 2022, Erasmus University Rotterdam
Quantib sponsors thesis-defenses on occasion. The following summarized review concerns the research Martijn Starmans performed as part of his PhD on Quantitative imaging biomarker development1.
Context
Healthcare is shifting from a uniform approach towards a more proactive, personalized approach based on the unique characteristics of each patient. Biomarkers, which are technologies that leverage big data to relate specific patient characteristics to clinical variables, are often used in the personalized clinical decision process. Especially in cancer medicine, there is a high need for accurate biomarkers, as cancer is a heterogeneous disease with a wide variety of manifestations.
As such, medical imaging such as CT and MRI is playing an increasingly important role. To create biomarkers from these images, the field of radiomics uses computer algorithms2 to extract quantitative, objective measurements from these images and relate them to biological processes, clinical outcomes, or diseases. Usually, hundreds or thousands of features are extracted from the data, which may be predefined or learned by the algorithm itself. A feature can contain any type of information useful for the task at hand. For example, when looking at a tumor, we can look at its shape (e.g., circular, star-like), intensity (e.g., high, low) or texture (e.g., uniform, heterogeneous). Next, artificial intelligence is used to decide which features relate to the outcome of interest and how these can be optimally combined in a predictive model.
Radiomics has, in recent years, shown many successes in identifying and extracting such biomarkers. In various clinical applications, radiomics has been used to predict diagnosis, prognosis, therapy response, surgery success, follow-up, and so on. For every new clinical application, the main challenge is to find a suitable radiomics method from the wide variety of available options. Each method can have different steps, for which there are multiple possible algorithms with parameters that need to be tuned per application. As most steps are not independent, and the performance of an algorithm depends on its parameter values, finding the most suitable algorithm and parameter values for each step is not trivial.
Currently, this is done manually through a heuristic trial-and-error process. This process has several disadvantages, as it: is time-consuming; requires expert knowledge; does not guarantee that an optimal solution is found; negatively affects the reproducibility; has a high risk of overfitting when not carefully conducted, and limits the translation to clinical practice.
The field faces several additional challenges: there is a need for publicly sharing data to facilitate reproducibility, to develop accurate biomarkers, and to validate the performance and generalization of biomarkers; there is a lack of image acquisition standardization, and there is a lack of reproducibility of both radiomics methods and biomarkers. Overcoming these barriers is vital for the translation of radiomics models to clinical practice.
What did the researchers do?
In the thesis, my colleagues and I at the Erasmus University Rotterdam aimed to streamline radiomics research, facilitate its reproducibility, and simplify its application. To this end, we developed an adaptive radiomics framework3, which automatically finds the optimal combination of radiomics algorithms and parameters from a wide range of available options. We hypothesized that, instead of manually tuning a radiomics method per application, it should be possible to create one radiomics method that works on multiple applications. Clinically, radiomics applications may be independent and show substantial differences (e.g., prostate cancer versus Alzheimer’s disease). Technically, however, the radiomics methods used often show substantial overlap.
To this end, we stated radiomics as a modular workflow, i.e., a specific combination of various algorithms and their associated hyperparameters. Examples of components included in this workflow are feature extraction, feature selection, dimensionality reduction, dataset resampling, and machine learning. For each component, the framework includes a large collection of commonly used algorithms in radiomics. To automatically construct and optimize the complete radiomics workflow from this search space of algorithms and hyperparameters, we exploited recent advances in automated machine learning. Instead of selecting the single best workflow, hyperensembles were introduced to combine different workflows into a single model, improving both the performance of the resulting radiomics model and the stability of the workflow optimization.
We evaluated the use of our framework in twelve different, independent clinical applications to evaluate the generalizability. To maximize the clinical relevance, we focused on oncology applications with a clear need for clinical decision support systems. Moreover, we aimed to facilitate generalization of the resulting biomarkers across image acquisition protocols and thus across clinical centers, therefore increasing the feasibility of applying such a biomarker in routine clinical practice.
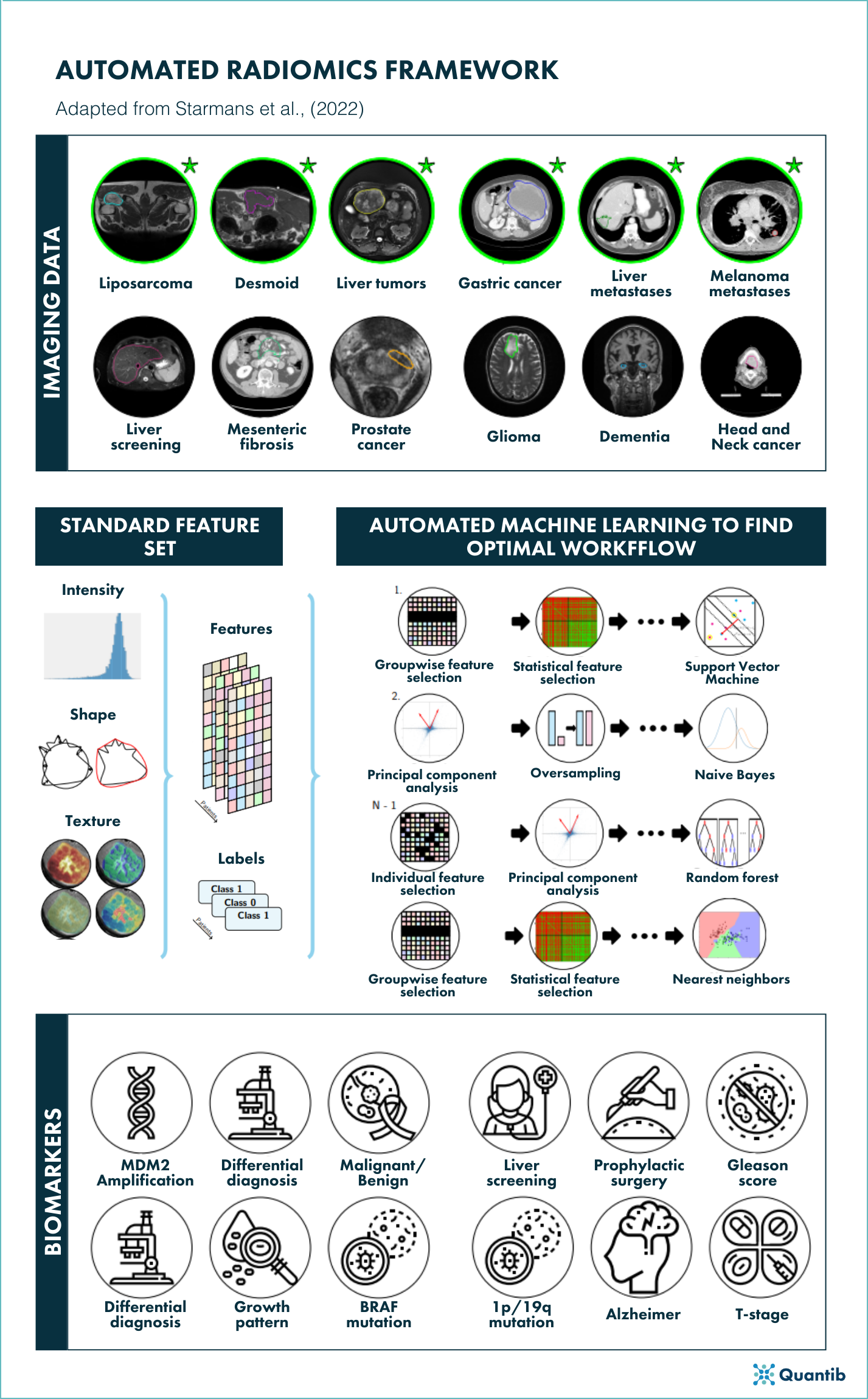
Figure 1. Schematic overview of the autimatic radiomics framework adapted from figure 13.1, Starmans, M., (2022).
What did the authors find?
The adaptive radiomics framework was evaluated in depth on twelve clinical applications. As an example, we evaluated whether radiomics can be used to differentiate eight different types of soft-tissue tumors (e.g., liposarcomas, gastrointestinal stromal tumors, and desmoid-type fibromatosis4-6). Our results showed that there is a relationship between radiomics features and the differential diagnosis, and that the radiomics models based on these features performed similar or even outperformed radiologists. Hence, radiomics can be used to distinguish between these tumors types, and thus holds the potential as an objective tool to improve clinical practice. Have a look at my thesis for many more examples.
To contribute to open-science, we have made both the software7 and a large database, consisting of data from 930 patients, publicly available8 for other researchers to use in order to reproduce our experiments or to train and validate their own radiomics models.
Conclusion
In this thesis, we addressed various of the challenges in the field of radiomics in order to streamline radiomics research, facilitate the reproducibility of radiomics methods and biomarkers, and ultimately simplify the use of radiomics in (new) clinical applications. To this end, we proposed a framework for to fully automatically optimize the construction of radiomics workflows to generalize radiomics across applications. The framework was evaluated on twelve different, independent clinical applications, on eleven of which our framework automatically constructed a predictive radiomics model. The adaptive framework can be used to efficiently probe datasets for biomarkers by conducting a validated, standardized radiomics baseline with minimal effort.
The resulting predictive models for the respective diseases have the potential to in the future improve the diagnostic trajectory and treatment planning of these patients. However, they are not yet ready for usage in clinical practice as they will have to be thoroughly tested by clinical research.
Bibliography
- Starmans MPA (2022) Streamlined Quantitative Imaging Biomarker Development: Generalization of radiomics through automated machine learning. Erasmus University Rotterdam, PhD Thesis. Available via http://hdl.handle.net/1765/137089
- Starmans MPA, van der Voort SR, Castillo T. JM, Veenland JF, Klein S, Niessen WJ (2020) Radiomics: Data mining using quantitative medical image features. In: Zhou SK, Rueckert D, Fichtinger G, (eds) Handbook of Medical Image Computing and Computer Assisted Intervention. Academic Press, pp 429-456. doi:10.1016/B978-0-12-816176-0.00023-5
- Starmans MPA, van der Voort SR, Phil T et al (2021) Reproducible radiomics through automated machine learning validated on twelve clinical applications. arXiv:210808618. arXiv:2108.08618. doi:arXiv:2108.08618
- Vos M, Starmans MPA, Timbergen MJM et al (2019) Radiomics approach to distinguish between well differentiated liposarcomas and lipomas on MRI. British Journal of Surgery 106:1800-1809. doi:10.1002/bjs.11410
- Timbergen MJM, Starmans MPA, Padmos GA et al (2020) Differential diagnosis and mutation stratification of desmoid-type fibromatosis on MRI using radiomics. European Journal of Radiology 131:109266. doi:10.1016/j.ejrad.2020.109266
- Starmans MPA, Timbergen MJM, Vos M et al (2022) Differential Diagnosis and Molecular Stratification of Gastrointestinal Stromal Tumors on CT Images Using a Radiomics Approach. Journal of Digital Imaging. 10.1007/s10278-022-00590-2. doi:10.1007/s10278-022-00590-2
- Starmans MPA, Van der Voort SR, Phil T, Klein S (2018) Workflow for Optimal Radiomics Classification (WORC). Zenodo, Available via https://github.com/MStarmans91/WORC. Accessed 22-12-2021. 10.5281/zenodo.3840534. doi:10.5281/zenodo.3840534
- Starmans MPA, Timbergen MJM, Vos M et al (2021) The WORC* database: MRI and CT scans, segmentations, and clinical labels for 930 patients from six radiomics studies. medRxiv. 10.1101/2021.08.19.21262238:2021.2008.2019.21262238. doi:10.1101/2021.08.19.21262238